Data Types
Babelomics has internally defined data types. Each file you upload to Babelomics or you generate using the software is tagged with one or more reserved words identifying such data types.
Data types are useful to avoid computation mistakes and also to help you doing a meaningful analysis.
You cannot use all data files within all Babelomics tools. Only certain data types can be selected from within each tool, those for which the tool or method has been devised for.
Babelomics defines and uses different data type for internal usage and validation:
- Microarray raw data
- Expresion data matrix
- ID lists
- Newick format
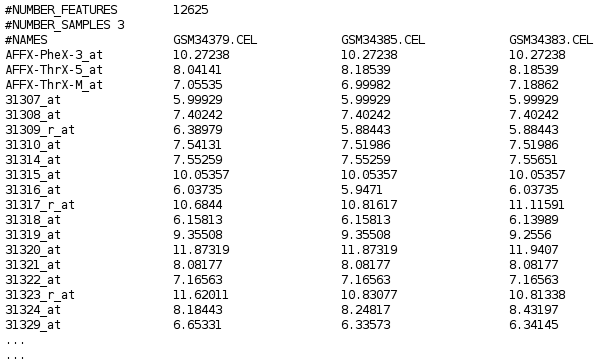
Expression data matrix
Expression matrices are numerical structures used to store expression data for many genomic features (genes, transcripts, exons…) form several samples (usually microarrays).
Babelomics arranges features in rows and samples in columns.
The first column of the matrix contains a name or ID for the genomic feature in each row. The first row of the file contains a name or ID for the sample.
These values are usually stored in tab delimited text files; meaning that the columns of the file are separated by the TAB character.
In the upper left top corner of the matrix the tag #NAMES is used by Babelomics to indicate that IDs are present in the matrix.
In the firs rows of the file there may be some comment lines starting by #.
An example of expression matrix file will look like this: