Table of Contents
2.1 Primary Analysis - Variants comparison
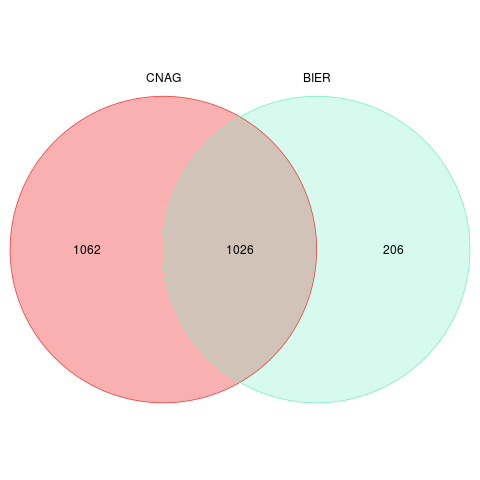
Overview
All unique variants detected
Unique variants detected in neuropathies-related genes
Summary by sample
- N_var: number of total variants detected by each pipeline
- N_var_neuropathies: number of variants detected by each pipeline found in neuropathies-related genes
- N_var intersect: number of total variants detected by both pipelines
- N_var_neuropathies intersect: number of variants detected by both pipelines and localised in neuropathies-related genes
| Sample | N_var_BIER | N_var_neuropathies BIER | N_var_CNAG | N_var_neuropathies CNAG | N_var intersect | N_var_neuropathies intersect |
|---|---|---|---|---|---|---|
| SGT038 | 53851 | 506 | 85942 | 902 | 42631 | 442 |
| SGT077 | 53230 | 487 | 83416 | 830 | 42191 | 415 |
| SGT161 | 52870 | 489 | 83533 | 866 | 42016 | 424 |
| SGT187 | 52320 | 539 | 84015 | 920 | 41339 | 459 |
| SGT230 | 52009 | 513 | 80023 | 854 | 41064 | 433 |
| SGT238 | 52960 | 526 | 82898 | 903 | 42230 | 466 |
| SGT241 | 52484 | 507 | 79876 | 863 | 41801 | 451 |
| SGT274 | 54182 | 473 | 84176 | 817 | 42885 | 410 |
Variants metrics by sample
- N_total variants: number of total variants detected by each pipeline
- N_hom: number of homozygous variants (1/1) detected by each pipeline
- N_het: number of heterozygous variants (0/1) detected by each pipeline
- N_biallelic: number of variants with one alternative allele (0/1 or 1/1) detected by each pipeline
- N_multiallelic: number of variants with more than one alternative allele (1/2) detected by BIER's pipeline
- N_SNPs: number of Single Nucleotide Polymorphisms detected by each pipeline
- N_indels: number of insertions or deletions detected by each pipeline
- N_Ts: number of variants involving transitions (A↔G, C↔T) detected by each pipeline
- N_Tv: number of variants involving transversions (A↔C, G↔T) detected by each pipeline
- Ts_Tv Ratio: ratio between the number of transitions and transversions
- %_PASS: percentage of variants tagged as “PASS” according to the parameters criteria assumed by each pipeline
- Mean quality: mean quality of variants detected by each pipeline
BIER's pipeline
| Sample | N_total variants | N_hom | N_het | N_biallelic | N_multiallelic | N_SNPs | N_indels | N_Ts | N_Tv | Ts_Tv Ratio | %_PASS | Mean quality |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SGT038 | 53851 | 19109 | 34686 | 53795 | 56 | 50431 | 3420 | 35362 | 15110 | 2.34 | 94.58 | 120283.15 |
| SGT077 | 53270 | 19025 | 34208 | 53233 | 37 | 50049 | 3221 | 35075 | 14995 | 2.34 | 95.22 | 573.19 |
| SGT161 | 52870 | 19235 | 33592 | 52827 | 43 | 49720 | 3150 | 34861 | 14899 | 2.34 | 94.92 | 590.80 |
| SGT187 | 52320 | 19779 | 32500 | 52279 | 41 | 49080 | 3240 | 34375 | 14731 | 2.33 | 94.68 | 41671.02 |
| SGT230 | 52009 | 20555 | 31414 | 51969 | 40 | 48668 | 3341 | 33942 | 14748 | 2.30 | 94.41 | 676.35 |
| SGT238 | 52960 | 19927 | 32995 | 52922 | 38 | 49652 | 3308 | 34881 | 14798 | 2.36 | 94.95 | 632.60 |
| SGT241 | 52484 | 19679 | 32767 | 52446 | 38 | 49234 | 3250 | 34591 | 14665 | 2.36 | 94.75 | 41551.44 |
| SGT274 | 54182 | 18994 | 35141 | 54135 | 47 | 50730 | 3452 | 35491 | 15273 | 2.32 | 94.54 | 79919.49 |
CNAG's pipeline
| Sample | N_total variants | N_hom | N_het | N_biallelic | N_SNPs | N_indels | N_Ts | N_Tv | Ts_Tv Ratio | %_PASS | Mean quality |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SGT038 | 85942 | 33194 | 52748 | 85942 | 80248 | 5694 | 57382 | 22866 | 2,51 | 82,27 | 168,38 |
| SGT077 | 83416 | 31780 | 51636 | 83416 | 78111 | 5305 | 55679 | 22432 | 2,48 | 80,69 | 164,92 |
| SGT161 | 83533 | 32702 | 50831 | 83533 | 78187 | 5346 | 55675 | 22512 | 2,47 | 81,22 | 166,04 |
| SGT187 | 84015 | 34875 | 49140 | 84015 | 78448 | 5567 | 55978 | 22470 | 2,49 | 81,47 | 167,02 |
| SGT230 | 80023 | 34525 | 45498 | 80023 | 74659 | 5364 | 53070 | 21589 | 2,46 | 82,54 | 171,34 |
| SGT238 | 82898 | 33457 | 49441 | 82898 | 77418 | 5480 | 55229 | 22189 | 2,49 | 81,94 | 168,63 |
| SGT241 | 79876 | 32252 | 47624 | 79876 | 74681 | 5195 | 53395 | 21286 | 2,51 | 82,12 | 170,69 |
| SGT274 | 84176 | 31984 | 52192 | 84176 | 78619 | 5557 | 55984 | 22635 | 2,47 | 82,21 | 168,49 |