Table of Contents
Exercise 5
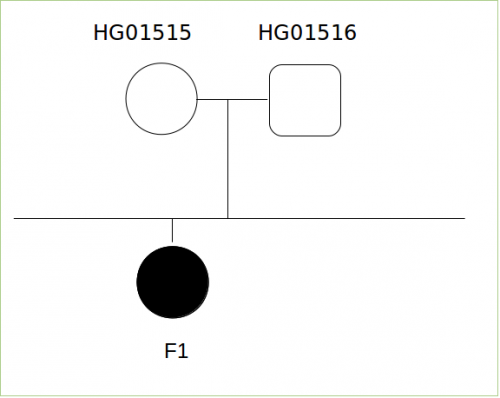
Data description
Working plan
We have several candidate profiles of mutations to explore:
How many variants do you detect for each scenario?
A. Individual filters
- Recessive heritage
- Dominant heritage (mother is affected).
- For chromosomes 1 and 3
- Evaluate these regions at the same time: 1:10000-500000, 11:1000-500000, 13:500-100000
- For theses genes: BRCA1,BRCA2
- Are there variants for these SNPs: rs6682385,rs75478250,rs147502335,rs368140013,rs7951297?
- Description of variants. How many SNVs, INDELs, MNVs, SVs, CNVs?
- Variants with MAF (Minimum Allelic Frequency) < 0.005 for Asian population in 1000 Genomes phase 1
- Variants with MAF (Minimum Allelic Frequency) < 0.005 for East Asian population in 1000 Genomes phase 3
- Variants with MAF (Minimum Allelic Frequency) < 0.005 for South Asian population in 1000 Genomes phase 3
B. Progressive selection
- There are clear information about our candidate variants:
- Recessive heritage
- Chromosomes 5 and 7
- INDEL
- How many variants do you have including both characteristics?
- Download these final results in a csv file
- We have other interesting scenario where we are sure there are several candidate variants:
- Heterozygous variant for mother and daughter and father is reference (0/0)
- Consequence type: “stop lost”
- We search new variants without rs ID
- BiERapp includes details about “Genomic context”, “Pop. Frequencies”, “Phenotype” and “Effect”. Please, could you explain a bit this information?