Table of Contents
Exercise 7
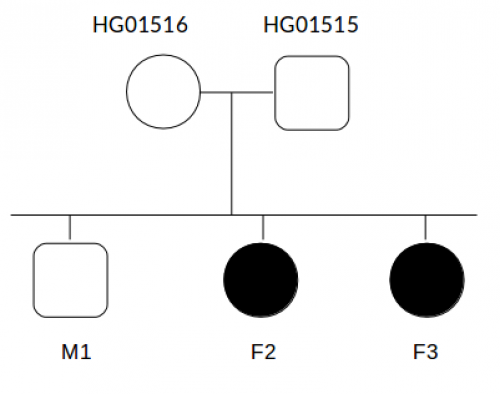
Data description
Working plan
We have several candidate profiles of mutations to explore:
How many variants do you detect for each scenario?
A. Individual filters
- Description of variants. How many SNVs, INDELs, MNVs, SVs, CNVs?
- Recessive heritage
- Dominant heritage (mother is affected).
- For this region: 3:10000-500000
- Evaluate these regions at the same time: 3:10000-500000, 5:1000-500000, 13:500-100000
- For theses genes related to Charcot-Marie-Tooth disease: BSCL2, DNM2, EGR2, FGD4, FIG4, GARS, GDAP1, GJB1, HSPB1, HSPB8, KIF1B, LITAF, LMNA, MFN2, MPZ, MTMR2, NDRG1, NEFL, PMP22, PRPS1, PRX, RAB7A, SBF2, SH3TC2, TRPV4, and YARS
B. Progressive selection
- We have several clues about our candidate variants:
- Recessive heritage
- For theses genes related to Charcot-Marie-Tooth disease: BSCL2, DNM2, EGR2, FGD4, FIG4, GARS, GDAP1, GJB1, HSPB1, HSPB8, KIF1B, LITAF, LMNA, MFN2, MPZ, MTMR2, NDRG1, NEFL, PMP22, PRPS1, PRX, RAB7A, SBF2, SH3TC2, TRPV4, and YARS
- How many variants do you have including both characteristics? Only one!
- Maybe this is our variant. We need several more things:
- Select this variant and take a picture from the Genomic Browser.
- Explore with detail information about “Genomic context”, “Pop. Frequencies”, “Phenotype”, “Effect”.
- Another interesting profile for us:
- Dominant heritage (mother is affected) where mother and daughters present a heterozygous variant.
- Only chromosome 5.
- How many variants do you have including both characteristics?
- Last scenario to explore:
- Recessive heritage but…..
- There are two affected daughters but one of them (F2) is more affected than (F3). Maybe there is a modifier for F2 not present in F3.
- Only chromosome 5.