Table of Contents
Data description
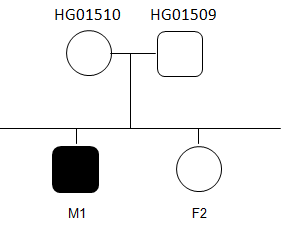
Working plan
If we have a clear profile of mutation, we will have to apply these filters on BiERapp to get directly the possible causal mutation. Sometimes we have several interesting profiles of mutations, not only one. For this case we should explore these different scenarios and decide the best prioritization strategy.
To know better this web tool, we propose you two groups of activities:
- In the part A of this activity, we would have to apply individual filters. This action give us information about the effect of each filter (maybe there are some filters more powerful than other). After each filtering, we have to clear the filter to apply the next one.
- In the part B of this exercise, we have a real selection where we combine several filters at the same time. It is interesting to know how the number of variants is reducing, when applying a progressive group of filters (without deleting the previous filter). We want to know the accumulative effect of several filters (all together).
How many variants do you detect for each scenario?
A. Individual filters
- Total number of variants without filters
- Recessive heritage
- Dominant heritage (mother and daughter are affected).
- For this region, only chromosome: 7
- Evaluate the same region, including start and end: 7:1000-200000
- For these genes: PMS,ATRN
- Are there variants for these SNPs: rs104895545,rs79716074?
- Description of variants. How many SNVs, INDELs, MNVs, SVs, CNVs?
- Variants with MAF (Minimum Allelic Frequency) < 0.001 for all populations in 1000 Genomes
- Variants with MAF (Minimum Allelic Frequency) < 0.001 for American populations in 1000 Genomes
- Variants with MAF (Minimum Allelic Frequency) < 0.001 for European-American population in ESP 6500
- Variants with the last two MAFs at the same time
- What do you thing about the effect of these filters? Between segregation and MAF, what is the most effective?
B. Progressive selection
- We have several clues about our candidate variants. In addition of knowing the pattern of recessive heritage, we search variants with MAF < 0.001 (for EXAC populations) because it is a rare disease.
- How many variants do you have including both characteristics?
- Include at the preview filter, the consequence type could be missense_variant. How many variants do you have now?
- Other scenario of interest:
- Candidate variants with dominant heritage (mother and daughter are affected).
- MAF < 0.001 for EXAC populations because it is a rare disease.
- Finally, include the SIFT < 0.05 and Polyphen > 0.95, thus it is likely pathogenic variant. How many variants do you have now?
- Download these final results in a csv file and check this file!
Solutions
Some of these solutions could be different when having updated databases
A. Individual filters
- Candidate variants: 39999
- Candidate variants: 1247
- Candidate variants: 2333
- Candidate variants: 2108
- Candidate variants: 1
- Candidate variants: 10
- Candidate variants: 0
- 36394 SNVs, 3582 INDELs, 19 MNVs, 4 SVs, 0 CNVs
- Candidate variants for all populations in 1000 Genomes: 700
- Candidate variants for American population in 1000 Genomes: 386
- Candidate variants for European-American population in ESP 6500 : 19728
- Candidate variants in the two last MAFs: 342
B. Progressive selection
- Candidate variants: 1247 → 518 → 1
- Candidate variants: 2333 → 993 → 7