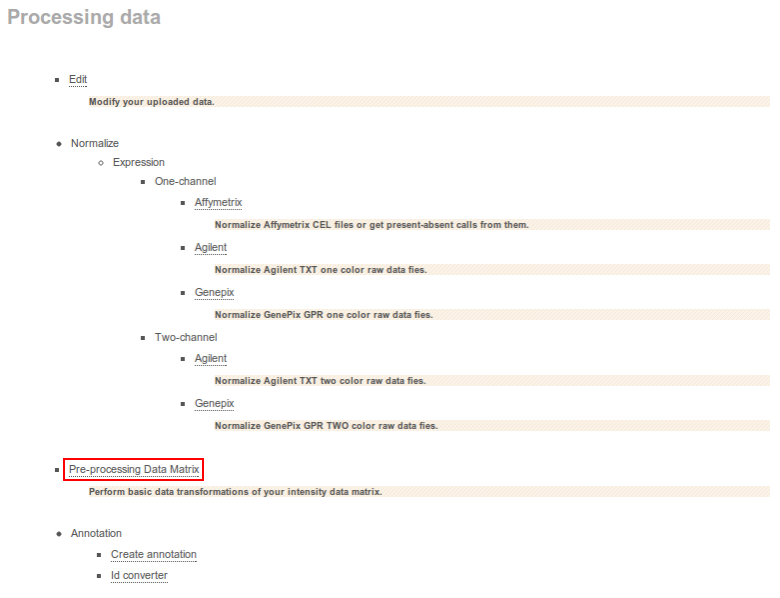
Pre-processing Data Matrix
With this tool you will be able to…
- Go to Processing data and then select Pre-processing Data Matrix.
- You will be redirected to a Pre-processing data matrix form. Here you can:
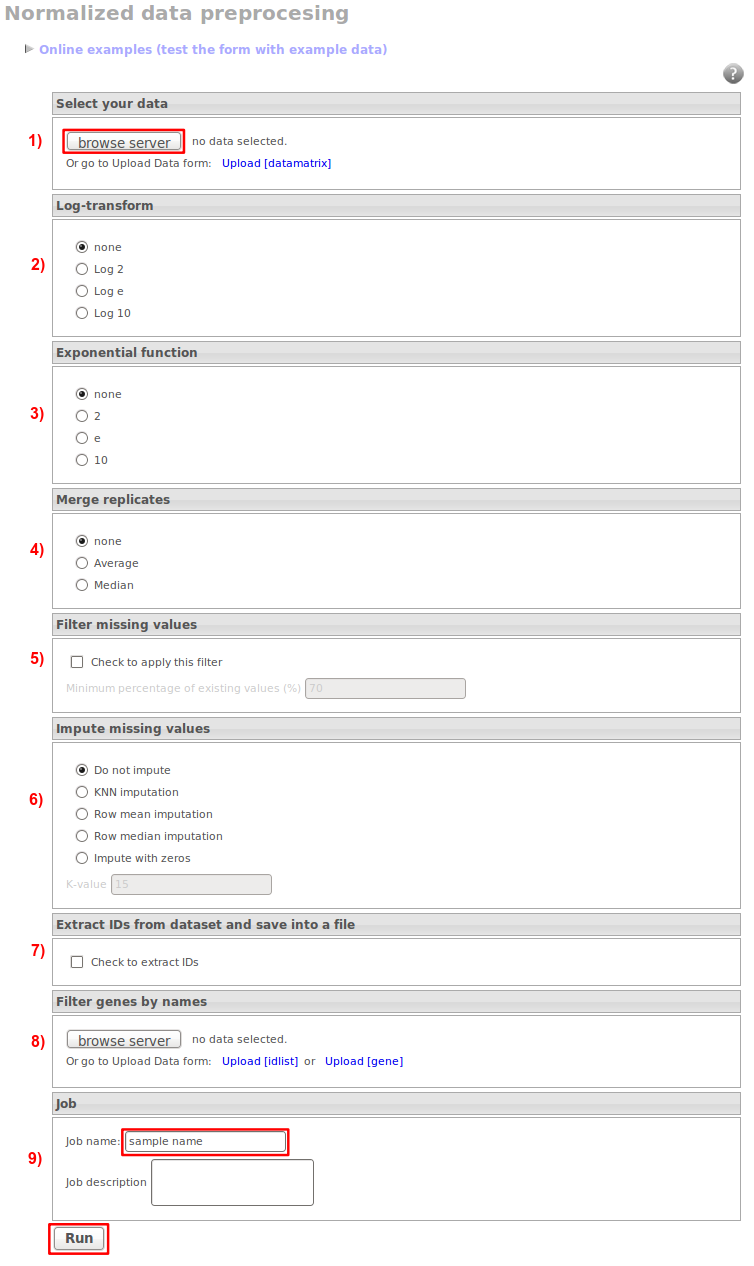
1) Select a data matrix of your interest. If it is not in your stored data, upload it.
2) Log transformation
This function calculates the logarithm of the expression values. You can select the base you prefer for this.
3) Exponential function
4) Merge replicates
This function looks for replicated clones (ids, genes…) and merge their patterns. You can choose between averaging the original patterns or getting the median.
5) Filter missing values
This option is intended for removing the patterns with many missing values. You can choose the 'Minimum percentage of existing values' you want to impose.
For example, if you have a dataset with 10 conditions and you set up the minimum percentage of existing values to 70%, all the patterns with less than 7 existing values will be removed, i.e., all the patterns with more than 3 missing values will be removed.
6) Impute missing values
This function fills out missing values. Several algorithms are available:
- fill with zeros: replace missing values by zeros. This is the simplest option and we do not recommend to use it unless you really know what you are doing.
- fill with row average: replace missing values by the row average. This option is better than the first one but again we do not recommend to use it unless you really know what you are doing.
- fill with row median: replace missing values by the. row median. This option is better than the first one but again we do not recommend to use it unless you really know what you are doing.
- KNNimpute: replace missing values by the average value of the K nearest patterns. You need at least 5 non-mising values for imputing the rest of the pattern. Good values for K are around 15.
See Troyanskaya et al. (2001) Missing value estimation methods for DNA microarrays. Bioinformatics 17 (6), pp. 520-525
7) Extract IDs from dataset and save into a file
All the ID's will be saved in a single column into a .txt file.
8) Filter genes by names
This option will remove all the genes that are present in the extra list you upload.
Further information: