Table of Contents
Analysis of Whole-Exome Sequencing data for diagnosis of Inherited Peripheral Neuropathies
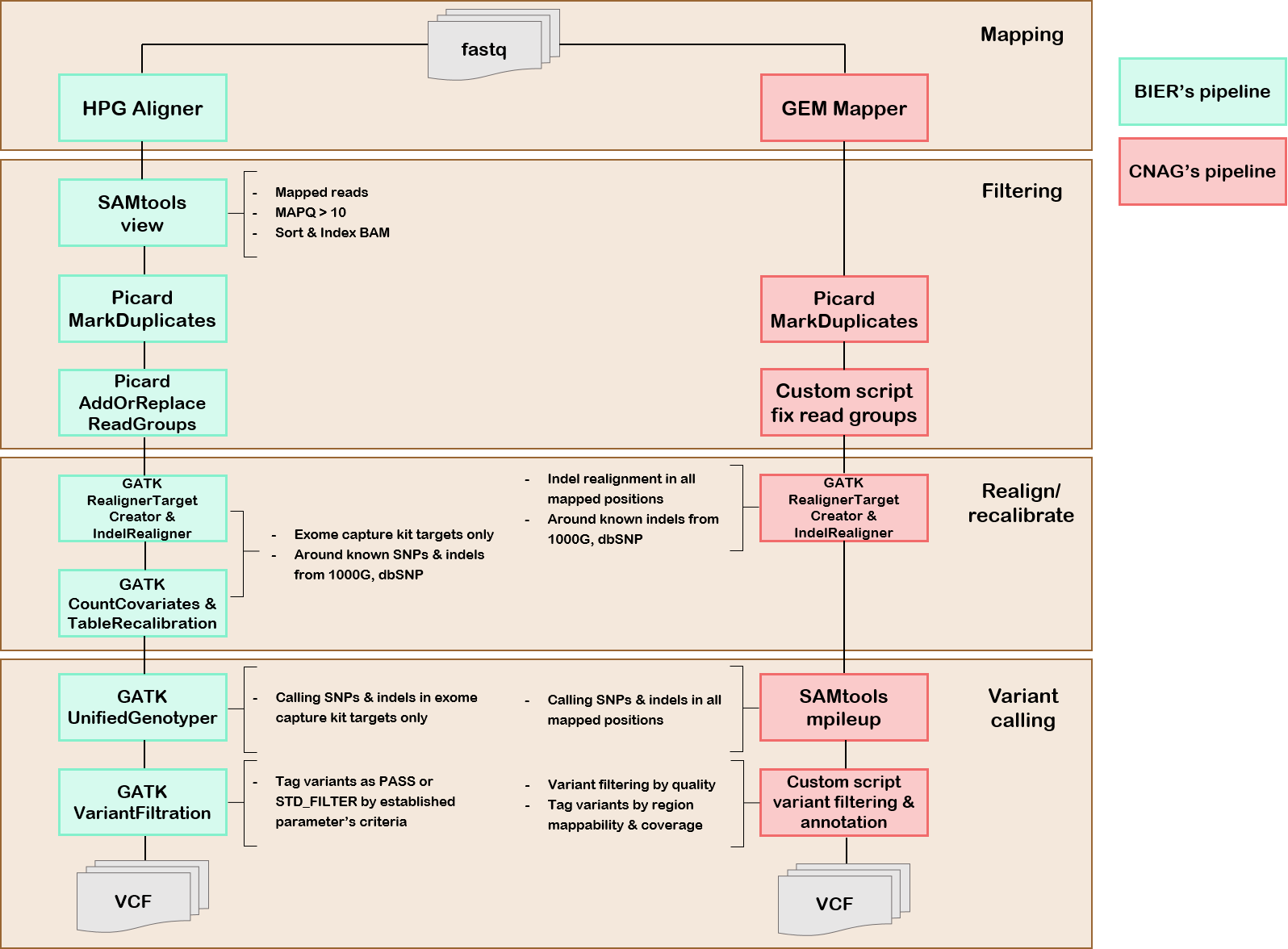
1. Methods
1.1 Pipeline's description
1.2 Primary Analysis
- For variants comparison, VCF files were manipulated using bash scripting to:
- Count total number of variants in whole exome and in neuropathies-related genes.
- Obtain a list of matching genomic positions in which variants had been found by both pipelines.
- Statistical metrics reports over variants were generated using VARIANT.
1.3 Coverage Analysis
- Sequencing coverage was evaluated in final BAM files from which variants were called in both pipelines using GATK DepthOfCoverage.
- For both pipelines, sequencing coverage was assessed in exome capture kit targets only. BEDtools was used to intersect a list of significant neuropathies-related genes genomic coordinates with a BED file containing exome capture kit targets (SureSelect Human All Exon V5) provided by the manufacturer.
- R scripts were used to manipulate coverage data generated by GATK DepthOfCoverage to calculate stats and build box and bar plots with ggplot2 package.