Get an experimental DataSet
The very first step is to get an experimental dataset. If we do not have our own one, we can easily find one in a public repository like GEO or ArrayExpress. They provide a user-friendly interface to query easily their databases. Both archives allow the user to browse or query the experiments via free text search (e.g. experiment accession numbers, authors, laboratory, publication, key words), and filter the experiments retrieved by species or array design or experiment type. Once the desired experiment is identified, the user can find more information about the samples, protocols used, experimental design, etc. and most importantly can export the data associated with the selected experiment.
The way of accessing to these repositories is described below.
Getting data from GEO
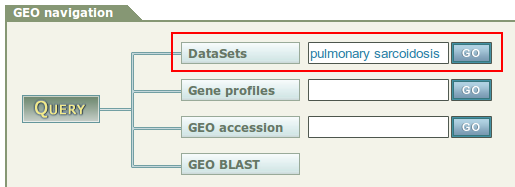
1. Go to the GEO home page: http://www.ncbi.nlm.nih.gov/geo/ GEO data can be retrieved in several ways:
2. Enter a keyword or any valid accessing code.
GEO data can be retrieved in several ways:
- To look at a particular GEO record for which you have the accession number, use the GEO accession box on the GEO homepage. (e.g: GSE16538)
- The simplest first step to find data relevant to your interests is to search Entrez GEO DataSets or Entrez GEO Profiles with keywords:
- Entrez GEO DataSets queries all experiment descriptions, allowing identification of studies of interest
- Entrez GEO Profiles queries gene expression profiles, allowing identification of genes of interest.
As with any other Entrez database, keywords or a simple Boolean phrase may be entered and restricted to any number of supported attribute fields, enabling effective query and mining of GEO data. Tools available under the 'Preview/Index' tab can help you construct complex, fielded queries.
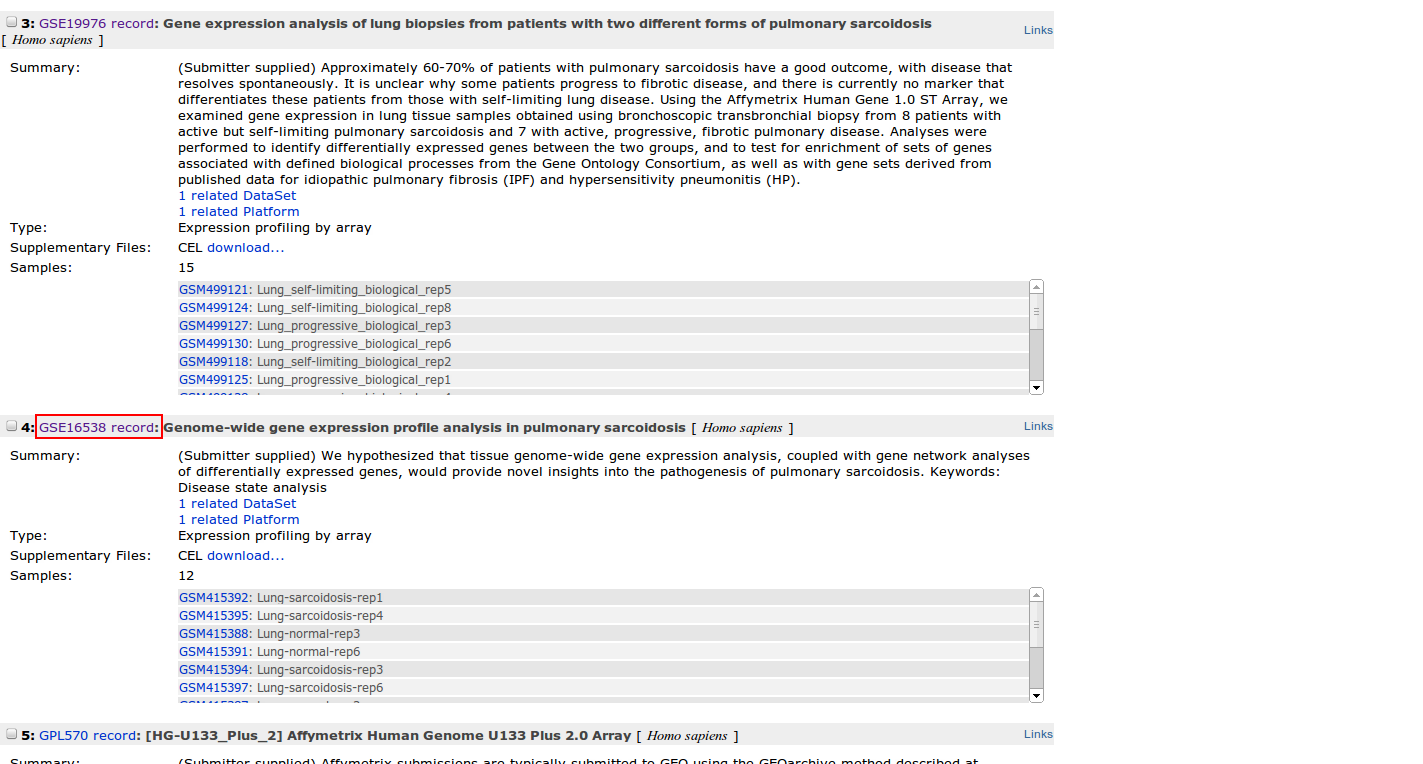
3. Identify a DataSet of interest
After querying GEO, we will get a list of results with the related DataSets. There are some features that will help us to identify the appropriate dataset:
- Summary: a few words about the analysis carried out with these samples.
- Organism: The specie analyzed.
- Type of experiment
- Subsets: The experimental groups contained in the DataSet.
- Samples: The number of samples and also the number of samples per subset.
4. Once you have identified a DataSet of interest, click on the record link. By accessing to this link we are redirected to a page with information about the experiment carried out (summary, sample description, etc.) and also about the authors and the PubmedID. We are going to focus our attention on information concerning the microarray chip and the samples. We can see that they have 12 samples (6 cases and 6 controls) and that the platform used is Affymetrix Human Genome U133 Plus 2.0 Array1).
In order to download the the raw data of the experiment, go to the bottom of the page and click on your favorite download mode: ftp or html.
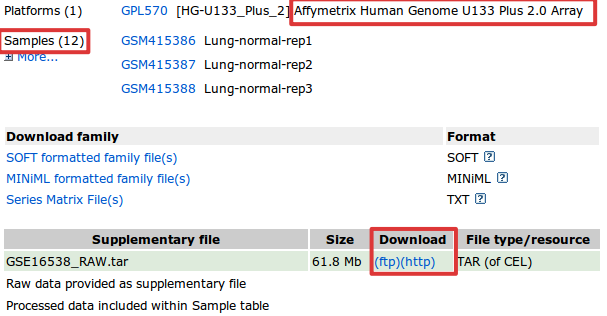
- The file downloaded is a compressed .tarfile and contains the necessary CEL files to go on with our study. If you are interested, you can uncompress the file and inside you will see other compressed files. Each file corresponds to a single sample.
Here you have an example:
| Archive/File | Name | Date | Time | Size | Type |
|---|---|---|---|---|---|
| Archive | GSE16538_RAW.tar | 06/11/2009 | 07:35:06 | 64798720 | TAR |
| File | GSM415386.CEL.gz | 06/10/2009 | 10:45:28 | 5516509 | CEL |
| File | GSM415387.CEL.gz | 06/10/2009 | 10:45:32 | 5514041 | CEL |
| File | GSM415388.CEL.gz | 06/10/2009 | 10:45:35 | 5396385 | CEL |
| File | GSM415389.CEL.gz | 06/10/2009 | 10:45:38 | 5391068 | CEL |
| File | GSM415390.CEL.gz | 06/10/2009 | 10:45:41 | 5321878 | CEL |
| File | GSM415391.CEL.gz | 06/10/2009 | 10:45:44 | 5370707 | CEL |
| File | GSM415392.CEL.gz | 06/10/2009 | 10:45:47 | 5273116 | CEL |
| File | GSM415393.CEL.gz | 06/10/2009 | 10:45:50 | 5347133 | CEL |
| File | GSM415394.CEL.gz | 06/10/2009 | 10:45:53 | 5442786 | CEL |
| File | GSM415395.CEL.gz | 06/10/2009 | 10:45:56 | 5474703 | CEL |
| File | GSM415396.CEL.gz | 06/10/2009 | 10:45:59 | 5400721 | CEL |
| File | GSM415397.CEL.gz | 06/10/2009 | 10:46:02 | 5329862 | CEL |
Getting data from ArrayExpress
1. Go to the ArrayExpress main homepage, at http://www.ebi.ac.uk/arrayexpress/
2. In the Experiments box, on the left-hand side of the page, type in a word or a phrase or GO term by which you want to retrieve the experiments, (e.g. 'stress') and click Query button.
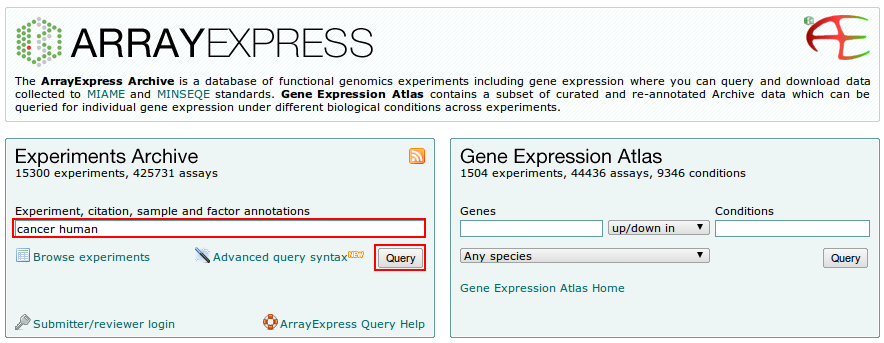
3. Choosing a DataSet.
This will bring up a window with a list of experiments in the reverse order of their publication. For each experiment the following information are displayed:
- Experiment accession number (ID): This is a unique identifier assigned to each experiment by the AE curation staff. The accession number can also be used to query the Archive.
- Title: with a brief description of the experiment.
- Number of assays associated with the experiment.
- Data availability: as processed or raw data.
By clicking the + button on the left-hand side of each row you will get a more detailed view of each experiment.
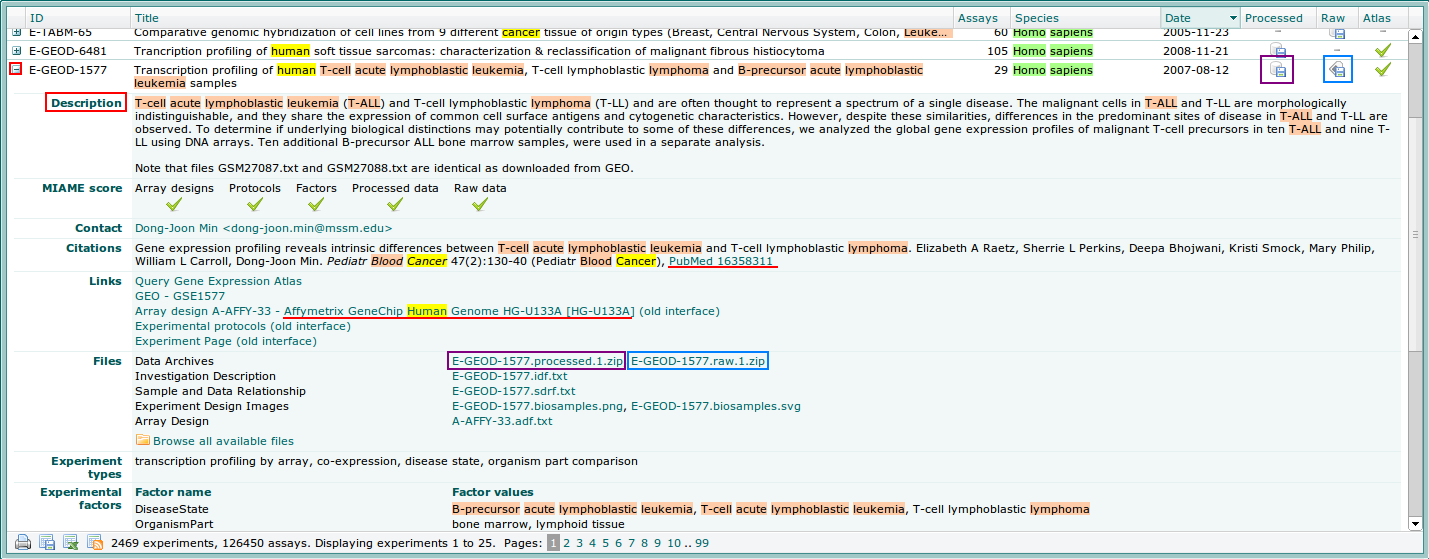
4. Downloading data.
Data is sometimes offered in two ways:
- Processed file: Already preprocessed and normalized. The downloaded file is a data matrix with the p-values for each sample and gene. Purple squares show the links to download processed data.
- Raw data file: .zip file with the CEL files related to this experiment containing the raw data for every feature on the chip. Blue squares show the links to download raw data.
- Affymetrix raw data files: .CEL files. (Affymetrix are always one color arrays).
- Agilent one color raw data files: one channel Agilent .TXT files.
- Agilent two color raw data files: two channel Agilent .TXT files.