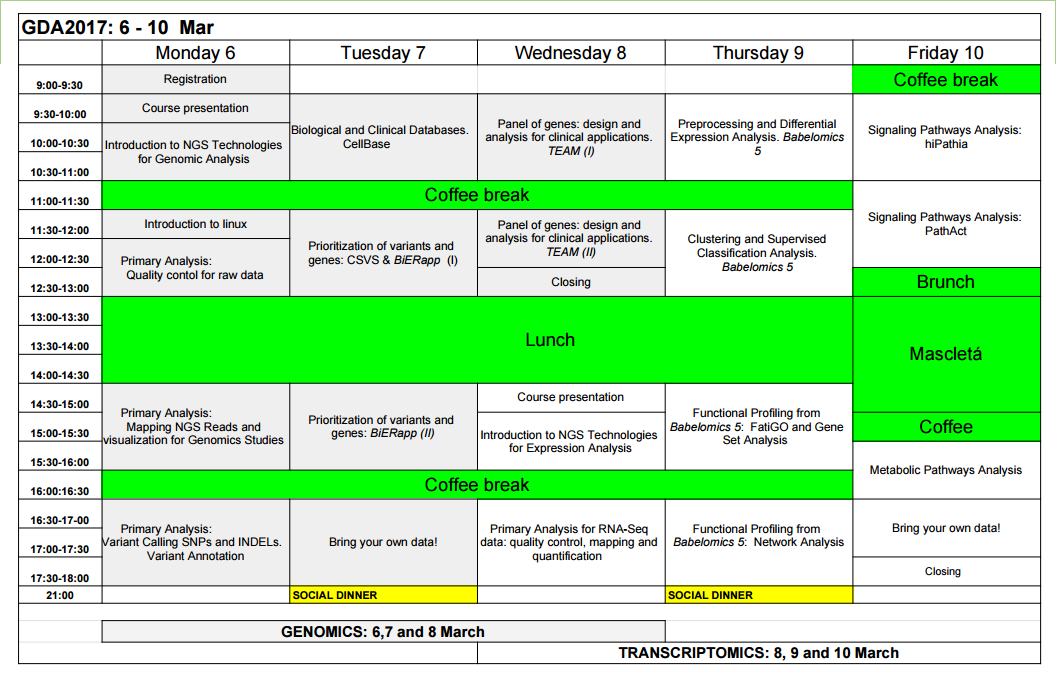
Table of Contents
GENOMIC VARIATION COURSE
Day 1
9:00 – 9:30 Registration
9:30 – 10:00 Course presentation
Joaquín Dopazo
- General overview of the course.
- NGS technologies, analysis methods and webtools.
10:00 – 11:00 Introduction to NGS technologies for Genomic Analysis
Joaquín Panadero
- Basics of the NGS technologies.
- Computing infrastructure for NGS analyses.
- Some remarks on experiment design.
11:00 – 11:30 Coffee break
11:30 – 12:00 Introduction to Linux
Francisco Salavert
12:00 – 13:00 Quality Control for Raw Data
Matías Marín
13:00 – 14:30 Lunch break
Will be held in the cafeteria of the CIPF.
14:30 – 16:00 Mapping NGS Reads and Visualization for Genomic Studies
Daniel Crespo
16:00 – 16:30 Coffee break
16:30 – 18:00 Variant Calling SNPs and INDELs. Variant Annotations
José Carbonell
Day 2
9:30 – 11:00 Biological and Clinical Databases
Alicia Amadoz
11:00 – 11:30 Coffee break
11:30 – 13:00 Prioritization of variants and genes: BiERapp (I) & CSVS
Francisco García, Francisco Salavert
13:00 – 14:30 Lunch break
Will be held in the cafeteria of the CIPF.
14:30 – 16:00 Prioritization of variants and genes: BiERapp (II)
Francisco García
16:00 – 16:30 Coffee break
16:30 – 18:00 Bring your own data!
Matías Marín, Daniel Crespo, Francisco García, Francisco Salavert, José Carbonell, Alicia Amadoz, Susi Gallego
- During this session you can analyze your data with us.
- You can also do the exercises of other sessions.
- If you do not have any data, this will not be a problem and here we have a full practice for you: NGS tutorial.
21:00 Social dinner
Day 3
9:30 – 11:00 Panel of Genes: Design and Analysis for Clinical Applications. TEAM (I)
Susi Gallego
TEAM (Targeted Enrichment Analysis and Management): Management of panels of genes for targeted enrichment and massive sequencing for diagnostic applications. How to design panels of genes.
11:00 – 11:30 Coffee break
11:30 – 12:30 Panel of Genes: Design and Analysis for Clinical Applications. TEAM (II)
Susi Gallego, Francisco García
12:30 – 13:00 Closing
Joaquín Dopazo
13:00 – 14:30 Lunch break
Will be held in the cafeteria of the CIPF
TRANSCRIPTOMICS COURSE
Day 1
14:30 – 15:00 Course presentation
Joaquín Dopazo
15:00 – 16:00 Introduction to NGS technologies for Expression Analysis
Joaquín Panadero
16:00 – 16:30 Coffee break
16:30 – 18:00 Primary Analysis for RNA-Seq Data: Quality Control, Mapping and Quantification
Daniel Crespo
Many colleagues had problems running tophat due to a problem with the installation of Bowtie2 in the virtual machine. To solve this, download the bash.bashrc file to your downloads folder and execute the following commands (password: bioclass):
- unzip ~/Downloads/bash.bashrc.zip
- sudo cp /etc/bash.bashrc /etc/orig.bash.bashrc
- sudo mv ~/Downloads/bash.bashrc /etc/
- sudo apt-get install bowtie2
Close the terminal and open a new one. tophat should work now.
Day 2
9:30 – 11:00 Normalization & Differential Expression Analysis
Matías Marín
11:00 – 11:30 Coffee break
11:30 – 13:00 Clustering and Supervised Classification Analysis
Cankut Çubuk
gda_clustering_and_prediction
GDA_clustering_and_prediction_exercises
prediction_clustering_exercises.zip
13:00 – 14:30 Lunch break
Will be held in the cafeteria of the CIPF.
14:30 – 16:00 Functional Profiling from Babelomics 5: FatiGO and Gene Set Analysis
Alicia Amadoz
16:00 – 16:30 Coffee break
16:30 – 18:00 Functional Profiling from Babelomics 5: Network Analysis
José Carbonell
21:00 Social dinner
Day 3
9:30 – 11:00 Signaling Pathways Analysis: hiPathia
Marta R. Hidalgo
11:00 – 12:30 Signaling Pathways Analysis: PathAct
Marta R. Hidalgo
12:30 – 13:00 Brunch
Will be held in the cafeteria of the CIPF.
13:00 – 15:00 Mascletà
15:00 – 15:30 Coffee break
15:30 – 16:30 Metabolic Pathways Analysis: Metabolizer
Cankut Çubuk
16:30 – 17:30 Bring your own data!
Matías Marín, Daniel Crespo, Francisco García, Francisco Salavert, José Carbonell, Alicia Amadoz, Susi Gallego, Cankut Çubuk, Marta Hidalgo
- During this session you can analyze your data with us.
- You can also do the exercises of other sessions.
17:30 – 18:00 Closing
References and links
- High throughput estimation of functional cell activities reveals disease mechanisms and predicts relevant clinical outcomes. Hidalgo MR, Cubuk C, Amadoz A, Salavert F, Carbonell-Caballero J, Dopazo J. Oncotarget. 2017;8(3):5160-5178
- Integrated gene set analysis for microRNA studies. Garcia-Garcia F, Panadero J, Dopazo J, Montaner D. Bioinformatics. 2016 Sep 15;32(18):2809-16. doi: 10.1093/bioinformatics/btw334. Epub 2016 Jun 20.PMID: 27324197
- Actionable pathways: interactive discovery of therapeutic targets using signaling pathway models. Salavert F, Hidago MR, Amadoz A, Çubuk C, Medina I, Crespo D, Carbonell-Caballero J, Dopazo J. Nucleic Acids Res. 2016
- Babelomics 5.0: functional interpretation for new generations of genomic data. Alonso R, Salavert F, Garcia-Garcia F, et al. Nucleic Acids Res. 2015;43:W117-W121.
- Assessing the impact of mutations found in next generation sequencing data over human signaling pathways. Hernansaiz-Ballesteros RD, Salavert F, Sebastián-León P, Alemán A, Medina I, Dopazo J. Nucleic Acids Res. 2015;43:W270-W275.
- Understanding disease mechanisms with models of signaling pathway activities. Sebastian-Leon P, Vidal E, Minguez P, et al. BMC Syst Biol. 2014;8(1):121.
- Inferring the functional effect of gene expression changes in signaling pathways. Sebastián-León P, Carbonell J, Salavert F, Sanchez R, Medina I, Dopazo J. Nucleic Acids Res. 2013;41:W213-W217.
- A web-based interactive framework to assist in the prioritization of disease candidate genes in whole-exome sequencing studies. Alemán A, Garcia-Garcia F, Salavert F, Medina I, Dopazo J. Nucleic Acids Res. 2014;42:W88-W93.
- A web tool for the design and management of panels of genes for targeted enrichment and massive sequencing for clinical applications. Alemán A, Garcia-Garcia F, Medina I, Dopazo J. Nucleic Acids Res. 2014;42:W83-W87.
- SNOW, a web-based tool for the statistical analysis of protein-protein interaction networks. Minguez P, Gotz S, Montaner D, Al-Shahrour F, Dopazo J. Nucl. Acids Res. 2009;37:W109-114
- Genome Maps, a new generation genome browser. Medina I, Salavert F, Sanchez R, et al. Nucleic Acids Res. 2013;41:W41-W46.