Command disabled: backlink
Input files
Expression data
There are two ways to upload your expression data in PATHiWAYS, compress raw data or a normalized matrix
- Raw data consists in a compress file containing a CEL file for each sample. PATHiWAYS tool allows both ZIP and TAR compression. You can download you data from a public database as Gene Expression Omnibus (GEO) (TAR compress file) or you can compress your CEL files in an unique compress files using ZIP or TAR compression.
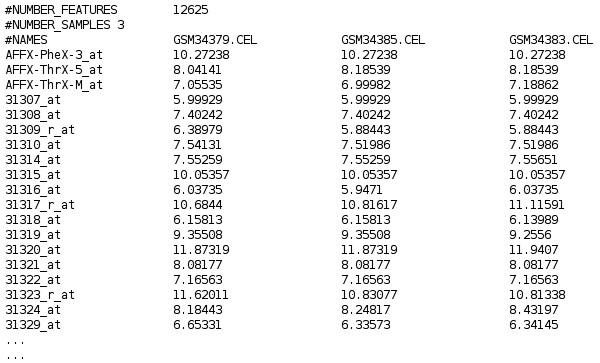
- Normalized expression matrices are numerical structures used to store normalized expression data for genomic features (in this case-probe sets in a determined Affymetrix microarray) from several samples. Probeset are in rows and features in columns. This normalization has to be done with rma method. You can use Babelomics in order to normalize correctly your CEL data. The first column of the matrix contains a name or ID for the genomic feature in each row. The first row of the file contains a name or ID for the sample. An example of expression matrix file will look like this:
Experimental design data
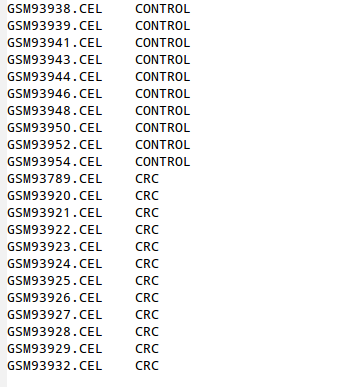
Is a tab delimited file with two columns.
The first one indicate the names of the samples indicated in the first row of the normalized matrix (if you choose to upload the normalized expression matrix) or the name of the CEL files (if you choose to upload raw data).
The second one indicate the experimental condition of each sample in the experiment.
It is possible to have more than two conditions in the second column but the analysis is possible only between two of these conditions.